Subscribe to and Analyze PlanetScope Imagery
This is a guide that will walk you through the process of subscribing to PlanetScope imagery for a set of agriculture fields. We will cover the following steps:
- Uploading areas of interest to feature collections
- Creating PlanetScope subscriptions
- Analyzing data to create indices and statistics
Prerequisites
- A Planet API key with access to an Area Under Management subscription
- A Sentinel Hub account
Set up your environment
from datetime import datetime, timezone, timedelta
from shapely.geometry import shape
from pprint import pprint
import geopandas as gpd
import pandas as pd
import requests
import numpy as np
import asyncio
import json
import copy
import os
import re
# Planet SDK
import planet
from planet.clients.subscriptions import SubscriptionsClient
from sentinelhub import (
CRS,
DataCollection,
Geometry,
SentinelHubStatistical,
SentinelHubStatisticalDownloadClient,
SHConfig,
SentinelHubRequest,
SentinelHubDownloadClient,
MimeType,
)
from ipyleaflet import Map, GeoData, basemaps, LayersControl
import matplotlib.pyplot as plt
import matplotlib.dates as mdates
%matplotlib inline
# Helper function to printformatted JSON using the json module
def p(data):
print(json.dumps(data, indent=2))
Upload areas of interest to feature collections
To order data, you need to provide areas of interest. You can do this by uploading GeoJSON features to a feature collection either through Features Manager or the Features API. Here we will use the Features API.
geojson_data = {"type":"FeatureCollection","features":[{"type":"Feature","properties":{},"geometry":{"coordinates":[[[-81.07147945222945,43.67490435965843],[-81.06495615860527,43.67223547577635],[-81.05882821611033,43.680122637871705],[-81.06521972602458,43.682624391132805],[-81.06785540021637,43.67912190735805],[-81.06663640090248,43.67864536315142],[-81.06920618323896,43.67585750370887],[-81.07026045291556,43.67631024182887],[-81.07147945222945,43.67490435965843]]],"type":"Polygon"}},{"type":"Feature","properties":{},"geometry":{"coordinates":[[[-81.05958597244029,43.675285619095064],[-81.05652200119249,43.67916956157043],[-81.05882821611033,43.68007498441628],[-81.06195807921277,43.67611961566857],[-81.05958597244029,43.675285619095064]]],"type":"Polygon"}},{"type":"Feature","properties":{},"geometry":{"coordinates":[[[-81.10129289138283,43.666119496077926],[-81.09704630815027,43.66439999452453],[-81.0921587312212,43.670698153259366],[-81.09645873059603,43.67237883850956],[-81.10129289138283,43.666119496077926]]],"type":"Polygon"}},{"type":"Feature","properties":{},"geometry":{"coordinates":[[[-81.05854129903942,43.6692934112491],[-81.06097545527373,43.6701737628008],[-81.06124321245954,43.66957512515077],[-81.06221687495348,43.66899408877978],[-81.06282541401194,43.66918776819534],[-81.06284975557425,43.669663160473675],[-81.06365302713158,43.67008572822684],[-81.06625757430251,43.66659945528957],[-81.06713387054724,43.66649380749428],[-81.06725557835875,43.66614164683307],[-81.06876475522422,43.66538449441549],[-81.0692515864712,43.66445124690017],[-81.06847265647616,43.66254949007825],[-81.06783977585489,43.662003604307216],[-81.06742596929533,43.66158097965604],[-81.06706084585984,43.66135205672751],[-81.06628191586482,43.661686636097585],[-81.06557601055715,43.66212686926988],[-81.06423722462823,43.662056432179355],[-81.05854129903942,43.6692934112491]]],"type":"Polygon"}}]}
# You may also choose to use your own geojson file for your own field boundaries
# Convert the GeoJSON data into a GeoDataFrame
features = [shape(feature["geometry"]) for feature in geojson_data["features"]]
properties = [feature["properties"] for feature in geojson_data["features"]]
agriculture_fields = gpd.GeoDataFrame(properties, geometry=features)
# Plotting setup
fig, ax = plt.subplots()
agriculture_fields.plot(ax=ax, color='#3366cc', edgecolor='black', alpha=0.6)
# Customize plot appearance
ax.set_title("Agriculture Fields")
ax.set_xlabel("Longitude")
ax.set_ylabel("Latitude")
# Show plot
plt.show()
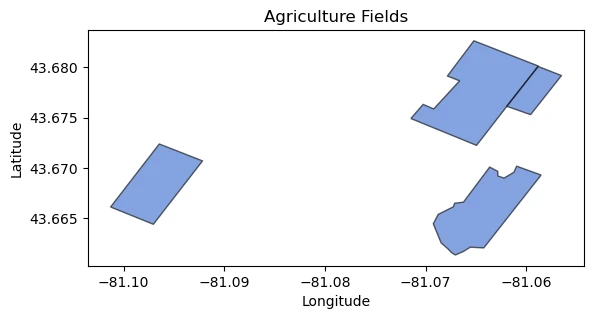
With your API key, you can connect the features API.
# Set up authentication
PL_API_KEY = os.getenv("PL_API_KEY")
# Set up Planet Features API base URL
URL = "https://api.planet.com/features/v0/ogc/my/collections"
# Set up the session
session = requests.Session()
session.auth = (PL_API_KEY, "")
Create a feature collection
Now, you will create a feature collection to store the uploaded features.
request = {
"title" : "Agriculture Fields",
"description" : "A collection of agriculture field boundaries."
}
# Send the POST request to the API stats endpoint
res = session.post(URL, json=request)
# Print response
p(res.json())
# let's save the collection-id to use later
feature_collection_id = res.json()["id"]
{
"id": "agriculture-fields-JvVxVDr",
"title": "Agriculture Fields",
"description": "A collection of agriculture field boundaries.",
"item_type": "feature",
"links": [
{
"href": "https://api.planet.com/features/v0/ogc/my/collections/agriculture-fields-JvVxVDr",
"rel": "self",
"title": "This collection",
"type": "application/json"
},
{
"href": "https://api.planet.com/features/v0/ogc/my/collections/agriculture-fields-JvVxVDr/items",
"rel": "features",
"title": "Features",
"type": "application/json"
}
],
"feature_count": 0,
"area": null,
"title_property": null,
"description_property": null,
"permissions": {
"can_write": true,
"shared": false,
"is_owner": true
},
"ownership": {
"owner_id": 359883,
"org_id": 1
}
}
Upload features to collection
With a feature collection created, you can upload features to the collection.
feature_refs = []
# Loop through each feature in the FeatureCollection
for i, feature in enumerate(geojson_data["features"]):
# Extract geometry from feature
geo = feature["geometry"]
# Build request data with unique ID and properties
request = {
"geometry": geo,
"id": str(i + 1), # Generate a unique ID for each feature, you can adjust this
"properties": {
"title": f"Feature-{i + 1}", # Customize title as needed
"description": f"Description for Feature-{i + 1}" # Customize description as needed
}
}
# Make the POST request
res = session.post(f"{URL}/{feature_collection_id}/items", json=request)
# Process response and get short reference
short_ref = res.json()[0] # Adjust depending on API response format
# Print or store the short reference for later use
print(f"Short reference for feature {i + 1}: {short_ref}")
feature_refs.append(short_ref)
Short reference for feature 1: pl:features/my/agriculture-fields-JvVxVDr/1-0aLYrRg
Short reference for feature 2: pl:features/my/agriculture-fields-JvVxVDr/2-mEW2XBg
Short reference for feature 3: pl:features/my/agriculture-fields-JvVxVDr/3-gBNDYVg
Short reference for feature 4: pl:features/my/agriculture-fields-JvVxVDr/4-o4wld7m
For each input feature, we now have a corresponding unique identifier, called a feature reference. You can use these feature references to create subscriptions which will deliver PlanetScope data to your imagery collections.
Create PlanetScope subscriptions
We will iterate over each feature in the feature collection and create a subscription for it. To do this, we will use the Planet Python SDK.
You will need to specifiy a start and end time, as well as the item and asset types you want to subscribe to. In this example, we will subscribe to PlanetScope 8-band imagery.
A delivery desintation is also required for subscriptions. In this example, data is delivered to an image collection on Planet Insights Platform. You can also deliver data to your own cloud storage destinations.
# Set up authentication
pl_api_key = os.getenv("PL_API_KEY")
auth = planet.Auth.from_key(pl_api_key)
# Set up variables for creating the subscriptions
# Define a start and end time
start_time = datetime(2024, 3, 1, tzinfo=timezone.utc)
end_time = datetime(2024, 11, 1, tzinfo=timezone.utc)
# Set a collection ID, or set it to None and the function will a create a new collection
collection_id = None # "insert-collection-id-for-pre-existing"
# List of subscriptions created
subscriptions = {}
async with planet.Session(auth=auth) as sess:
cl = SubscriptionsClient(sess)
#For each feature in our geojson feature collection
for feature_ref in feature_refs:
feature_collection_id = short_ref.split("/")[-2]
feature_id = short_ref.split("/")[-1]
# Create a name for the subscription
subscription_name = f"{feature_collection_id}_{feature_id}_PlanetScope"
# Build the subscription payload
payload = planet.subscription_request.build_request(
name=subscription_name,
source=planet.subscription_request.catalog_source(
start_time=start_time,
end_time=end_time,
item_types=["PSScene"],
asset_types=["ortho_analytic_8b_sr", "ortho_analytic_8b_xml", "ortho_udm2"],
geometry={
"type": "ref",
"content": feature_ref,
},
),
hosting=planet.subscription_request.sentinel_hub(collection_id=collection_id),
)
# Create the subscription
results = await cl.create_subscription(payload)
subscription_id = results["id"]
subscriptions[feature_ref] = subscription_id
print(f"Feature Reference: {feature_ref} -> Subscription ID: {subscription_id}")
# If no collection ID is set, set the collection ID as the collection created for the first subscription that is created
if not collection_id:
results = await cl.get_subscription(subscription_id=subscriptions[0])
collection_id = results["hosting"]["parameters"]["collection_id"]
await asyncio.sleep(3) # delay so collection can be established
print(f"{len(subscriptions)} Subscriptions created. Data is being delivered to the following collection: {collection_id}")
Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/1-0aLYrRg -> Subscription ID: 9242db6f-d325-4e4a-bfc2-be431025dd8d
Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/2-mEW2XBg -> Subscription ID: ef6c1d03-5965-4bfd-b4b4-4414f7dbe37b
Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/3-gBNDYVg -> Subscription ID: 9bc73504-d779-403e-b698-1fef8699c86b
Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/4-o4wld7m -> Subscription ID: 266d0642-14be-4c2b-9446-1900b79c6371
4 Subscriptions created. Data is being delivered to the following collection: 65834003-ca52-41b9-b089-34a8fa4eb9c0
Check subscription statuses
You now have one feature per area of interest and one subscription for each feature. You can poll the Subscriptions API to get their statuses.
async with planet.Session(auth=auth) as sess:
cl = SubscriptionsClient(sess)
for feature_ref, subscription_id in list(subscriptions.items()):
sub_details = await cl.get_subscription(subscription_id=subscription_id)
subscription_status = sub_details["status"]
print(f"({subscription_status}) Feature Reference: {feature_ref} -> Subscription ID: {subscription_id}")
(running) Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/1-0aLYrRg -> Subscription ID: 9242db6f-d325-4e4a-bfc2-be431025dd8d
(running) Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/2-mEW2XBg -> Subscription ID: ef6c1d03-5965-4bfd-b4b4-4414f7dbe37b
(running) Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/3-gBNDYVg -> Subscription ID: 9bc73504-d779-403e-b698-1fef8699c86b
(running) Feature Reference: pl:features/my/agriculture-fields-JvVxVDr/4-o4wld7m -> Subscription ID: 266d0642-14be-4c2b-9446-1900b79c6371
Delivering data will take several minutes - continue to check in on the status. As data is delivered, you can jump into the second section. Until data is delivered, the section section will not work.
Analyze data to create indices and statistics
Now that data has been delivered to an image collection, you can use the Statistical and Process APIs to analyze the PlanetScope imagery that was ordered.
You must first install and configure the sentinelhub-py package.
config = SHConfig()
if not config.sh_client_id or not config.sh_client_secret:
print("No credentials found, please provide the OAuth client ID and secret.")
else:
print("Connected")
Connected
The following code will fetch the features from the API, extract geometries and properties from the features, create a GeoDataFrame, and reproject it to EPSG:3857. It's optimal to use a projected coordinate system for inputs into these APIs when working with small areas. This allows you to specify a resolution at which to analyze the data in units of meters instead of degrees, which is useful for small areas.
# Fetch the features from the API
res = session.get(f"{URL}/{feature_collection_id}/items")
features = res.json()['features']
# Extract geometries and properties from features
geometries = [shape(feature["geometry"]) for feature in features]
properties = [feature["properties"] for feature in features]
# Create GeoDataFrame and reproject to EPSG:3857
agriculture_fields = gpd.GeoDataFrame(properties, geometry=geometries, crs="EPSG:4326")
agriculture_fields_3857 = agriculture_fields.to_crs(epsg=3857)
# Display the reprojected GeoDataFrame
agriculture_fields_3857
| title | description | pl:ref | pl:area | geometry | |
|---|---|---|---|---|---|
| 0 | Feature-4 | Description for Feature-4 | pl:features/my/agriculture-fields-JvVxVDr/4-o4... | 405934.196941 | POLYGON ((-9023395.542 5414406.396, -9023666.5... |
| 1 | Feature-3 | Description for Feature-3 | pl:features/my/agriculture-fields-JvVxVDr/3-gB... | 314480.137995 | POLYGON ((-9028154.627 5413917.953, -9027681.9... |
| 2 | Feature-2 | Description for Feature-2 | pl:features/my/agriculture-fields-JvVxVDr/2-mE... | 106278.548884 | POLYGON ((-9023511.834 5415328.625, -9023170.7... |
| 3 | Feature-1 | Description for Feature-1 | pl:features/my/agriculture-fields-JvVxVDr/1-0a... | 548812.596246 | POLYGON ((-9024835.810 5415269.945, -9024109.6... |
Create NDVI zonal statistics time series
For each area of interest, we can create a Statistical API request to calculate the NDVI zonal statistics time series. We will use the SentinelHubStatistical class from the sentinelhub-py package to create the requests.
You will need to provide inputs for
- source imagery collection
- time interval
- spatial resolution
- an evalscript
- specification for aggregation intervals and histograms for statistics
image_collection = DataCollection.define_byoc(collection_id)
input_data = SentinelHubStatistical.input_data(image_collection)
# Set a time interval
time_interval = start_time.strftime('%Y-%m-%d'), end_time.strftime('%Y-%m-%d')
# Specify a resolution
resx = 3
resy = 3
# Provide an evalscript
evalscript = '''//VERSION=3
function setup() {
return {
input: [
{
bands: [
"red",
"nir",
"dataMask",
"clear"
]
}
],
output: [
{
id: "ndvi",
bands: 2
},
{
id: "dataMask",
bands: 1
}
]
}
}
function evaluatePixel(samples) {
let ndvi = (samples.nir-samples.red)/(samples.nir+samples.red);
const indexVal = samples.dataMask === 1 ? ndvi : NaN;
let id_default = colorBlend(ndvi, [0.0, 0.5, 1.0],
[
[1,0,0, samples.dataMask * samples.clear],
[1,1,0,samples.dataMask * samples.clear],
[0.1,0.31,0,samples.dataMask * samples.clear],
])
return {
ndvi: [ndvi, samples.clear * samples.dataMask],
dataMask: [samples.dataMask],
};
}'''
# Create the requests
aggregation = SentinelHubStatistical.aggregation(
evalscript=evalscript, time_interval=time_interval, aggregation_interval="P1D", resolution=(resx, resy)
)
histogram_calculations = {"ndvi": {"histograms": {"default": {"nBins": 20, "lowEdge": -1.0, "highEdge": 1.0}}}}
# For each polygon field boundary, create a Statistical API request
agriculture_fields_3857 = agriculture_fields.to_crs(3857)
ndvi_requests = []
for geo_shape in agriculture_fields_3857.geometry.values:
request = SentinelHubStatistical(
aggregation=aggregation,
input_data=[input_data],
geometry=Geometry(geo_shape, crs=CRS(agriculture_fields_3857.crs)),
calculations=histogram_calculations,
config=config,
)
ndvi_requests.append(request)
print(f"{len(ndvi_requests)} Statistical API requests prepared")
4 Statistical API requests prepared
With the Statistical API requests prepared, you can download the data and create a time series of NDVI zonal statistics.
%%time
download_requests = [ndvi_request.download_list[0] for ndvi_request in ndvi_requests]
client = SentinelHubStatisticalDownloadClient(config=config)
ndvi_stats = client.download(download_requests)
print("{} Results from the Statistical API!".format(len(ndvi_requests)))
4 Results from the Statistical API!
CPU times: total: 1.59 s
Wall time: 41.8 s
Data is returned as json objects. We can normalize the data into a pandas DataFrame for further analysis.
ndvi_dfs = [pd.json_normalize(per_aoi_stats["data"]) for per_aoi_stats in ndvi_stats]
for df, feature_title in zip(ndvi_dfs, agriculture_fields.title):
df["feature_title"] = feature_title
ndvi_df = pd.concat(ndvi_dfs)
# calculate date as day of the year for time integration
ndvi_df["day_of_year"] = ndvi_df.apply(lambda row: datetime.fromisoformat(row['interval.from'].rstrip('Z')).timetuple().tm_yday, axis=1)
# create a date field as a date data type
ndvi_df["date"] = pd.to_datetime(ndvi_df['interval.from']).dt.date
# delete and drop unused columns
del_cols = [i for i in list(ndvi_df) if i not in ["interval.from", "outputs.ndvi.bands.B0.stats.mean", "outputs.ndvi.bands.B1.stats.mean", "day_of_year", "feature_title", "date"]]
ndvi_df = ndvi_df.drop(columns=del_cols).rename(columns={'interval_from': 'date', 'outputs.ndvi.bands.B0.stats.mean': 'ndvi_mean', 'outputs.ndvi.bands.B1.stats.mean': 'clear', 'feature_title':'feature_title'})
# assign datatypes to float columns
ndvi_df['ndvi_mean'] = ndvi_df['ndvi_mean'].astype(float)
ndvi_df['clear'] = ndvi_df['clear'].astype(float)
ndvi_df
| interval.from | ndvi_mean | clear | feature_title | day_of_year | date | |
|---|---|---|---|---|---|---|
| 0 | 2024-03-01T00:00:00Z | 0.014131 | 0.000000 | Feature-4 | 61 | 2024-03-01 |
| 1 | 2024-03-04T00:00:00Z | 0.354233 | 1.000000 | Feature-4 | 64 | 2024-03-04 |
| 2 | 2024-03-07T00:00:00Z | 0.374601 | 1.000000 | Feature-4 | 67 | 2024-03-07 |
| 3 | 2024-03-08T00:00:00Z | 0.102291 | 0.000000 | Feature-4 | 68 | 2024-03-08 |
| 4 | 2024-03-10T00:00:00Z | 0.027932 | 0.000000 | Feature-4 | 70 | 2024-03-10 |
| ... | ... | ... | ... | ... | ... | ... |
| 149 | 2024-10-24T00:00:00Z | 0.741877 | 0.653461 | Feature-1 | 298 | 2024-10-24 |
| 150 | 2024-10-26T00:00:00Z | 0.771047 | 1.000000 | Feature-1 | 300 | 2024-10-26 |
| 151 | 2024-10-27T00:00:00Z | 0.118916 | 0.000000 | Feature-1 | 301 | 2024-10-27 |
| 152 | 2024-10-28T00:00:00Z | 0.697495 | 1.000000 | Feature-1 | 302 | 2024-10-28 |
| 153 | 2024-10-30T00:00:00Z | 0.472776 | 0.128951 | Feature-1 | 304 | 2024-10-30 |
610 rows × 6 columns
# Suppress chained assignment warning for the specific line that requires it
pd.options.mode.chained_assignment = None
# Define the moving average function
def moving_average(x, w):
return np.convolve(x, np.ones(w), 'same') / w
# Initialize plot
fig, ax = plt.subplots(figsize=(13, 7))
# Set up date formatting for x-axis
ax.xaxis.set_major_formatter(mdates.DateFormatter('%Y-%m-%d'))
ax.xaxis.set_major_locator(mdates.WeekdayLocator(interval=10))
# Plot each fields rolling average NDVI
for idx, feature_title in enumerate(agriculture_fields.title):
series = ndvi_df[(ndvi_df["feature_title"] == feature_title) & (ndvi_df["clear"] >= 0.9)]
# Calculate rolling average and assign directly
series = series.copy() # |Prevent chained assignment warning
series["rolling_avg"] = moving_average(series["ndvi_mean"], 3)
# Plot rolling average
ax.plot(series["date"], series["rolling_avg"], label=f"Field {feature_title}", color=f"C{idx}")
# Set labels and title
ax.set_title('Mean NDVI Over Time', fontsize=32)
ax.set_ylabel("Mean NDVI", fontsize=28)
ax.set_xlabel("Date", fontsize=28)
ax.legend(title="Fields")
plt.show()
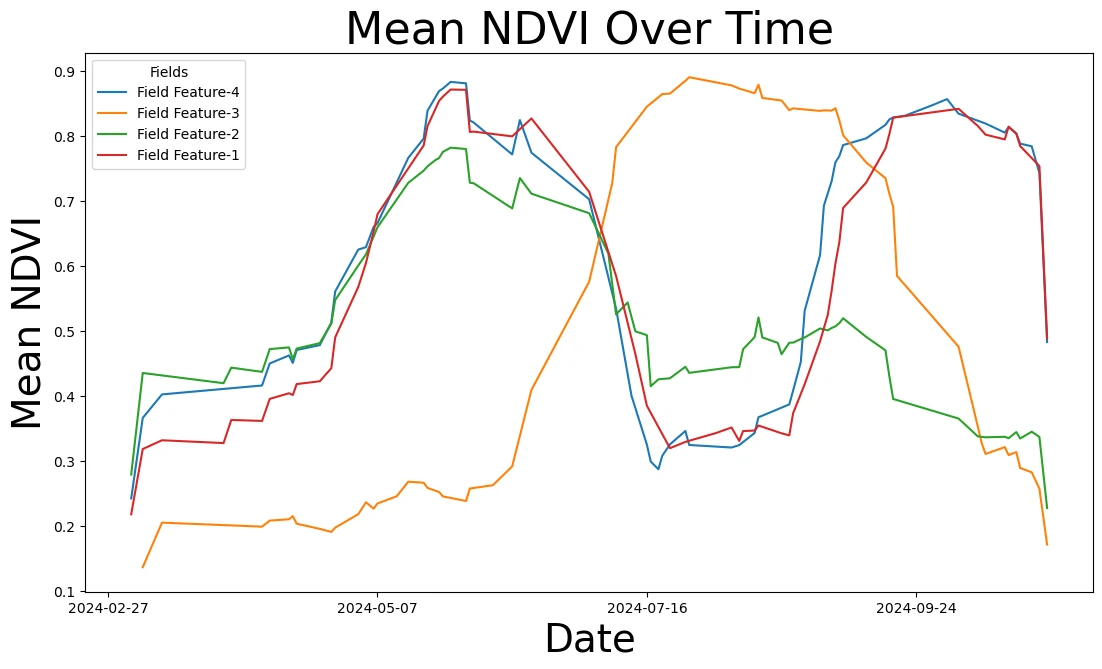
Create true color maps for cloud-free days
In this section, we will look at:
- Requesting true color imagery over cloud free days
- Performing multi-temporal analysis by caclulating the median NDVI from cloud-free images
To create our visualizations - let's start by selecting one field for the demonstration. In practice, you can batch these requests.
agriculture_field = agriculture_fields_3857.iloc[0]
print(agriculture_field)
agriculture_field["geometry"]
title Feature-4
description Description for Feature-4
pl:ref pl:features/my/agriculture-fields-JvVxVDr/4-o4...
pl:area 405934.196941
geometry POLYGON ((-9023395.541961536 5414406.39573724,...
Name: 0, dtype: object
# Let's grab certain dates to visualize with the Processing API
# Subset the time series to one field where it's >90% clear
one_field = ndvi_df[(ndvi_df["feature_title"] == agriculture_field["title"]) & (ndvi_df["clear"].ge(0.9))]
# Find the date with the highest NDVI
peak_ndvi_date = one_field.loc[one_field['ndvi_mean'].idxmax(), 'date']
# Calculate a column for date difference from peak NDVI
one_field['date_diff'] = (one_field['date'] - peak_ndvi_date).abs()
# Find the 5 dates where we have clear imagery closest to the peak NDVI
one_field_sorted = one_field.sort_values(by='date_diff')
unique_acquisitions = one_field_sorted.head(5).sort_values(by="date")['date']
unique_acquisitions_list = unique_acquisitions.to_list()
unique_acquisitions_list
[datetime.date(2024, 5, 24),
datetime.date(2024, 5, 26),
datetime.date(2024, 5, 30),
datetime.date(2024, 5, 31),
datetime.date(2024, 6, 1)]
To visualize data in true color, you can use the following evalscript. This script will return true color imagery with a data mask to show cloud-free pixels.
true_color_evalscript = '''
//VERSION=3
//True Color
function setup() {
return {
input: ["blue", "green", "red", "dataMask", "clear"],
output: { bands: 3 }
};
}
function evaluatePixel(sample) {
return [sample.red / 3000, sample.green / 3000, sample.blue / 3000, sample.dataMask*sample.clear];
}
'''
And we can then loop through each timestamp and create a Process API requests for each date.
process_requests = []
time_difference = timedelta(hours=1)
for timestamp in unique_acquisitions_list:
request = SentinelHubRequest(
evalscript=true_color_evalscript,
input_data=[
SentinelHubRequest.input_data(
data_collection=image_collection,
time_interval=(timestamp, timestamp),
)
],
responses=[SentinelHubRequest.output_response("default", MimeType.PNG)],
geometry=Geometry(agriculture_field["geometry"], crs=CRS("EPSG:3857")),
resolution=(3, 3),
config=config,
)
process_requests.append(request)
print("{} Process API requests prepared.".format(len(process_requests)))
5 Process API requests prepared.
%%time
client = SentinelHubDownloadClient(config=config)
download_requests = [request.download_list[0] for request in process_requests]
data = client.download(download_requests)
CPU times: total: 1.94 s
Wall time: 2.45 s
ncols, nrows = 5, 1
fig, axis = plt.subplots(
ncols=ncols, nrows=nrows, figsize=(15, 5),
subplot_kw={"xticks": [], "yticks": [], "frame_on": False}
)
for idx, (image, timestamp) in enumerate(zip(data, unique_acquisitions_list)):
ax = axis[idx]
ax.imshow(np.clip(image * 2.5 / 255, 0, 1))
ax.set_title(timestamp.isoformat(), fontsize=10)
plt.tight_layout()
plt.show()
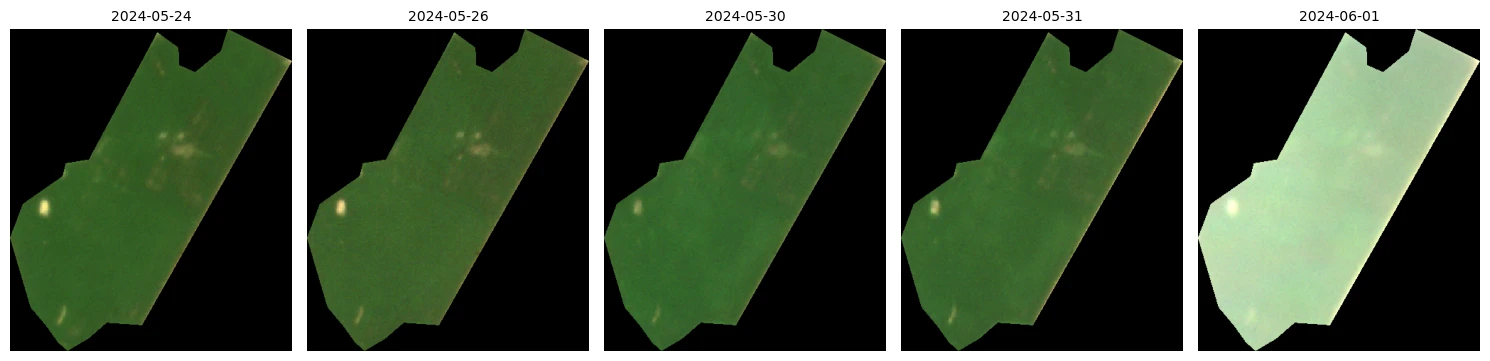
Create a median NDVI composite imagery with clouds removed
If you want to then take these multiple cloud free observations and create a composite for the median NDVI, you can do that with multitemporal analysis in the evalscript.
min_date = unique_acquisitions.min()
max_date = unique_acquisitions.max()
time_interval = min_date.strftime('%Y-%m-%d'), max_date.strftime('%Y-%m-%d')
print(time_interval)
('2024-05-24', '2024-06-01')
The following evalscript will calculate the median NDVI from the cloud-free images.
median_ndvi_evalscript = '''
function setup() {
return {
input: ["red", "nir", "dataMask", "clear"],
output: [{ id: "default", bands: 4 }],
mosaicking: "ORBIT" // Mosaicking method
};
}
// Function to get the median value from an array
function getMedian(values) {
values.sort((a, b) => a - b);
const middle = Math.floor(values.length / 2);
return values.length % 2 !== 0 ? values[middle] : (values[middle - 1] + values[middle]) / 2.0;
}
// Evaluate each pixel across all samples
function evaluatePixel(samples) {
let ndviValues = [];
for (let i = 0; i < samples.length; i++) {
let sample = samples[i];
// Include only cloud-free pixels (where clear and dataMask are both 1)
if (sample.dataMask === 1 && sample.clear === 1) {
let ndvi = index(sample.nir, sample.red);
ndviValues.push(ndvi);
}
}
// Calculate the median NDVI
let medianNDVI = ndviValues.length > 0 ? getMedian(ndviValues) : null;
return medianNDVI !== null
? colorBlend(medianNDVI,
[0.0, 0.5, 1.0],
[
[1, 0, 0, samples[0].dataMask],
[1, 1, 0, samples[0].dataMask],
[0.1, 0.31, 0, samples[0].dataMask],
])
: [0, 0, 0, 0]; // Transparent if no valid samples
}'''
request = SentinelHubRequest(
evalscript=median_ndvi_evalscript,
input_data=[
SentinelHubRequest.input_data(
data_collection=image_collection,
time_interval=time_interval,
)
],
responses=[SentinelHubRequest.output_response("default", MimeType.PNG)],
geometry=Geometry(agriculture_field["geometry"], crs=CRS("EPSG:3857")),
resolution=(3, 3),
config=config,
)
image = request.get_data()
plt.imshow(image[0])
plt.axis('off')
plt.title("Median NDVI Composite over 5 Days")
plt.show()
